

This year, the winners of MacHacks were none other than iBioMed Students, Isabelle Ragany and Daniel Duan, who collaborated with Aaron Li, a software engineering at Western.
MacHacks is one of Canada’s first major hackathons that is a competition that enables students from all disciplines to collaborate and produce innovative solutions. This year, the winners of MacHacks were none other than Isabelle Ragany (Level III – iBioMed and Electrical, McMaster), Daniel Duan (Level III – iBioMed and Electrical, McMaster), and Aaron Li (Level IV – Honours Spec. Computer Science w/ Minor in Software Engineering, Western). The iBioMed students were able to use their bioengineering backgrounds and as a team, extended their knowledge on the applications of enzymes with the use of their computer science skills to create “Denatured”.
To learn more about their Project “Denatured”, iBioMed conducted an interview with the winners of MacHacks:
Denaturing “Denatured”, break down what your winning project is about?
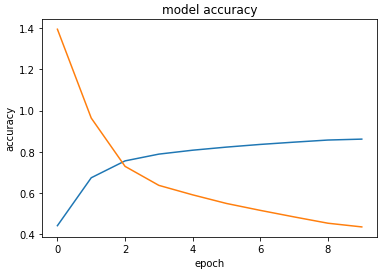
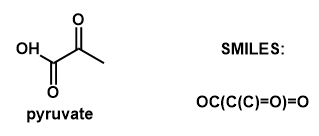
Denatured takes the reactants and products of a chemical reaction in the form of SMILES (a string of symbols that represent chemicals and their structures) and attempts to guess the Enzyme Commission number (EC #) required to complete the reaction. Enzyme Commission numbers are a method of classifying enzymes based on their function. Each number in the EC# of an enzyme describes a more specific class of enzymes that follow a similar function. In its most distilled form, a deep learning model can only take in numbers as inputs. However, our chemical reactions are not numerical. This means that we had to create a way of translating from characters to a numerical format all while preserving the features/information about the reaction. Once we have the numerical representation of our data, we can then give it to our neural network to allow it to train itself and find patterns within the data. Our network uses a special Long-Short Term Memory (LSTM) layer to process the sequential nature of the input data. Once the network learns the patterns in the data we give it, we can then try and give the network new reactions to try and guess the answers to based on previous patterns it has seen.
What did you enjoy most about this project?
We enjoyed learning and applying new knowledge in such a short period of time. None of our team members had previous experience in machine learning; so when our model showed promising results for the first time it was very rewarding! It was a great exercise in breaking down a problem into individual steps and tackling them one by one.
Were you able to apply the things you learned in iBioMed to this project?
The Python skills that we learned in iBioMed courses such as IBEHS 1P10, iBioMed’s first-year design course, came in handy! Since we used Google Colaboratory, the main language we worked in was Python. iBioMed’s interdisciplinary nature also exposed us to various chemistry and biology courses. This was helpful to get us started and figure out the key components to our design.
However, what was most useful was our previous experience in working in groups on projects. We were able to delegate tasks and coordinate our work efficiently. Additionally, the design courses in the iBioMed program helped us greatly! Each year in iBioMed there is a new design course for that year. These courses push us to learn and help us develop our research skills. This was an asset when working on a project where it was our first-time applying concepts such as natural language processing. In just 36 hours, we were able to learn and apply the tools needed to create our project.
Are you continuing to work on Denatured? If so, what’s next?
We would use a larger training dataset to cover more inputs and EC classifications. Our model was trained with a dataset of roughly 35000 cases. However, there are endless chemical reactions out there! Increasing our dataset to cover more inputs and subclasses of EC numbers would result in a better-trained model.
We are also considering different deep learning models to tackle the problem. A different approach would be to use a transformer, which is a newer deep learning model primarily used in the field of natural language processing. The transformer can weigh the significance of different parts of the data that is input. RNNs and transformers both handle sequential input data, however, in contrast to RNNs, transformers do not always process the data in order. This mechanism allows for relating different positions of a sequence to compute an overall representation of the sequence. This may help the model uncover additional patterns or features in reactions to better classify the EC number.
Is there anything else you would like people to know?
We all really enjoyed our experience at the MacHacks 2 hackathon! Hackathons like these are invaluable experiences to take what you’ve learned in the classroom and apply the skills. It is really rewarding to see how much you can do in such a short amount of time. We all highly recommend attending a hackathon. Not only is it a chance to learn new and exciting skills, but it is also an opportunity to network and meet new people.
Secondly, we recommend those who are interested to check out machine learning for themselves! While some of the technical languages on it may seem complex or confusing, getting started on it is actually very approachable and does not require complex programming skills.
Team Members of “Denatured”, MacHack Winners
- Isabelle Ragany – Level III – iBioMed and Electrical, McMaster
- Daniel Duan – Level III – iBioMed and Electrical, McMaster
- Aaron Li – Level IV – Honours Spec. Computer Science w/ Minor in Software Engineering, Western


